FIGURE 3
Computer models of the GR DBD from NMR structure determination with putative flanking alpha helices attached docked at a 39bp sequence of MMTPRGR1. A composite of amino acid and codon alignments from figure 2c are highlighted on the protein and DNA respectively. All highlighted residues have a dot surface indicating the van der Waals surfaces of each atom in that residue. The DNA nucleotides are color coded: Ade = green, Thy = red, Gua = yellow and Cyt = blue. Color coding of amino acids is based on polarity: positively charged side chains = blue, negatively charged side chains = red, uncharged polar side chains = yellow and nonpolar side chains = purple. The protein is docked at a distance of about 10 angstroms from the DNA for visual clarity. Above the DNA model in figure 3a, is a schematic of the DNA sequence. Boxes and underlines/overlines are as in figure 2b. Methylation inhibition studies by others (14) are summarized withthe following symbols: Triangles show nucleotides where methylation inhibitsreceptor binding. The circles show nucleotides which, when methylated, do not inhibit receptor binding but cannot be methylated after receptor is bound. The square shows a guanine that is hypermethylated in the presence of bound receptor
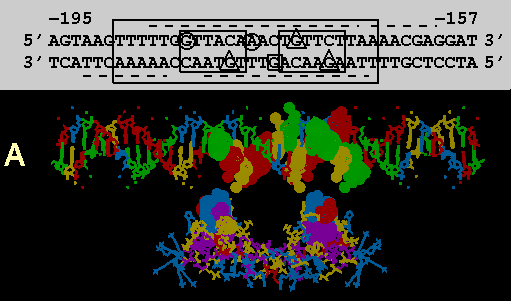
(a) Exon 3 encoded DNA recognition helix alignments
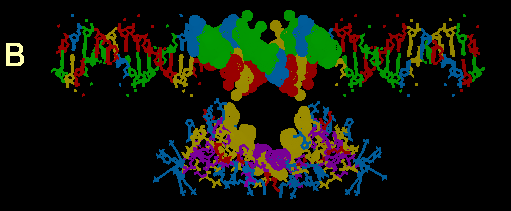
(b) Exon 4 encoded beta strand alignments.
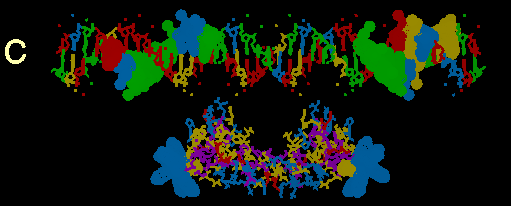
(c) Exon 5 encoded putative alpha helix alignments.