FIGURE 4
Plots of interaction energy between GRE nucleotides and selected GR DBD amino acids from the right monomer of the 10 Angstrom water layer GR DBD/29 bp GRE model after energy minimization, heating and equilibration. Amino acids are labeled using the Dayhoff one letter code and numbered as in the rat GR (48). Nucleotides are numbered as in figure2. Codon/anticodon nucleotides for the respective amino acids are boxed.
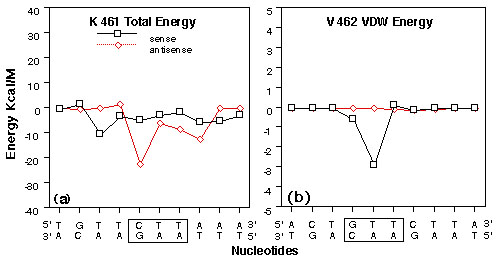
(a) Lysine 461 total interaction energies with GRE nucleotides 17-26 on the sense strand and 33-42 on the antisense strand.
(b) Valine 462 VDW interaction energies with GRE nucleotides 15-24 on the sense strand and 35-44 on the antisense strand.
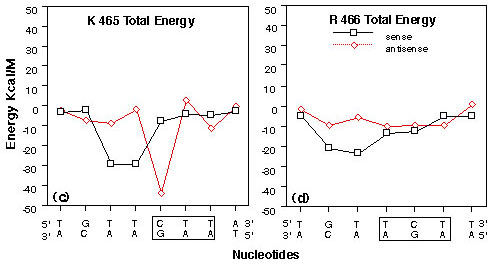
(c) Lysine 465 total interaction energies with GRE nucleotides 17-24 on the sense strand and 35-42 on the antisense strand.
(d) Arginine 466 total interaction energies with GRE nucleotides 17-23 on the sense strand and 36-42 on the antisense strand.
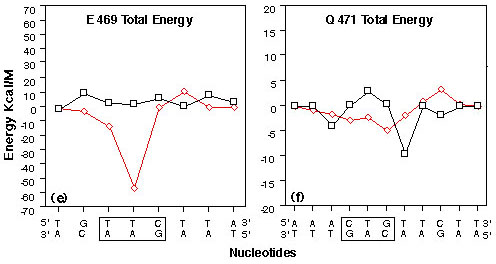
(e) Glutamic acid 469 total interaction energies with GRE nucleotides 17-24 on the sense strand and 35-42 on the antisense strand.
(f) Glutamine 471 total interaction energies with GRE nucleotides 13-23 on the sense strand and 36-46 on the antisense strand.