FIGURE 1
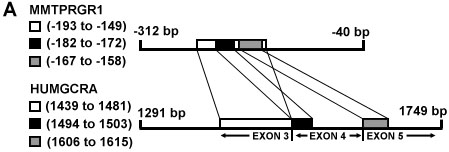

(a) A schematic of local nucleotide sequence alignments for exon 3: 1318 to 1485 bp, exon 4: 1486 to 1602 bp and exon 5: 1603 to 1626 bp of the GR DBD (GENBANK locus HUMGCRA) vs mouse mammary tumor virus 5' long terminal repeat (GENBANK locus MMTPRGR1) nucleotides ranging from -312 to -40 upstream from the MMTV transcription start site.
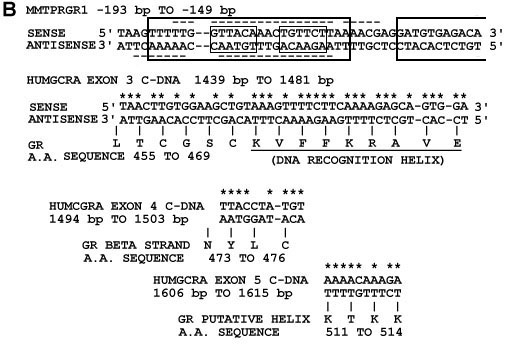
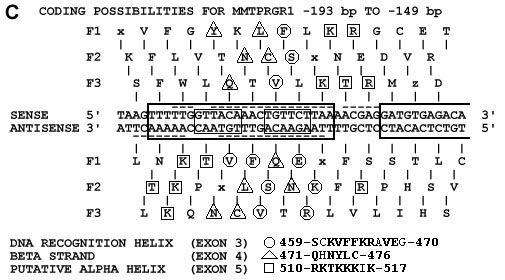
(b) Nucleotide sequence alignments from figure 1a. Above is shown the MMTPRGR1 nucleotide sequence within which GR binding sites have been detected with nuclease footprinting studies by others and are shown as large boxes (17) and dashed underlines and overlines (18-19). Small boxes contain the two glucocorticoid receptor binding half-sites GTTACA and TGTTCT respectively. Nuclecotide base pair matches are starred. Below the HUMGCRA cDNA sequences are shown their corresponding amino acid sequences in Dayhoff (47) one-letter code with the amino acids numbered as in the Rat GR (48). The recognition helix in the exon 3 alignment is underlined.